The intricate molecular processes underlying the cellular changes during plant immunity engage a high degree of proteomic plasticity and involve de novo synthesis of regulatory proteins, as well as their regulated degradation. Thus, it is inevitable to study the intimate interplay between protein synthesis, transport and degradation and how these pathways co-ordinate their functions to regulate plant immunity. In support of this, we discovered that regulated protein turnover via the ubiquitin-proteasome system (UPS) controls multiple aspects of plant immunity (Üstün et al., 2016). A significant role of the UPS in the regulation of plant immunity is further supported by the findings that plant pathogens have developed strategies to target and exploit the UPS for virulence. Several bacterial type-III effector proteins (T3Es) were shown to suppress plant defences by acting as E3 ligases (e.g. AvrPtoB), or by promoting ubiquitination of target proteins (e.g. HopM1). A more direct way to subvert the UPS is achieved by bacterial type-III effectors secreted that directly interact with proteasome components and thereby suppress proteasome function to suppress salicylic acid (SA)-mediated plant immunity (Üstün et al., 2013; Üstün et al., 2014; Üstün & Börnke, 2015). More recently, we revealed that Pst inhibits the proteasome by a certain effector repertoire, including HopM1 that seems to associate with UPS components (Üstün et al., 2016).
In addition to the UPS, autophagy is another major degradation pathway implicated mainly in cellular homeostasis, stress tolerance, and pathogen defence in eukaryotic organisms. Autophagic mechanisms rely on a core set of conserved AuTophaGy-related (ATG) genes to form double membrane vesicles, termed autophagosomes, that sequester and deliver cytoplasmic cargo to lytic compartments (i.e. lysosomes/vacuoles) for breakdown and recycling (Üstün et al., 2017). Increasing evidence now indicates that autophagy also acts as selective mechanism to degrade protein aggregates, organelles and specific proteins, e.g. in cellular quality control and under stress conditions. Selectivity is mediated by autophagy adaptors such as NBR1 (Neighbour of BRAC1), which binds to ubiquitylated substrates and autophagosome-associated ATG8 proteins. Importantly, recent findings revealed that the proteasome is eliminated by autophagy (“proteaphagy”) in response to chemical proteasome inhibition and nutrient limitation, suggesting a functional link between the two proteolytic systems in plants. Recently, we have identified that plant pathogenic bacterium Pseudomonas syringae pv. tomato DC3000 (Pst) activates autophagy and leads to the degradation of the proteasome by the proteaphagy pathway to enhance its pathogenicity (Üstün et al., 2018).
Based on our finding we suggest that plant pathogens target the two major proteolytic degradation pathways to have a broad impact on the plant side, possibly affecting the triangle of protein synthesis-transport-degradation. Pathogen-induced activation of autophagy and suppression of proteasome function will lead to a massive degradation and/or accumulation of targets implicated in defence and other cellular processes. Additional effects of autophagy activation and proteasome suppression on processes such as protein secretion and translation are currently unknown and will be investigated in the Üstün lab. Another goal of the Üstün lab is to understand how the action of T3Es is able to modulate proteolytic pathways. Based on preliminary and previous results, we will utilize effector proteins as tools to dissect the autophagy pathway to reveal novel regulators of autophagy.
Proteomic analysis of possible targets of pathogen-induced autophagy led to the discovery that in addition to the proteasome, other pathways such as RNA metabolism, protein translation and vesicle trafficking might be autophagy targets or regulators. Based on this preliminary data we have identified a vesicle trafficking component as a novel modulator of autophagy (Gouguet et al., unpublished). It appears that the loss of this and other trafficking components and interference with protein secretion at ER exit sites modulates autophagy responses revealing a novel layer of crosstalk between the endomembrane system and autophagy mechanism.
Using Pseudomonas and Xanthomonas effectors we have systematically screened them for their ability to interfere with proteasome function and autophagy. IP-MS experiments with candidate effector proteins revealed a subset of effectors interacting with autophagy components. So far, we have identified a Xanthomonas effector that is able to target an autophagy component to modulate autophagic turnover (Leong et al., in preparation), which we will further characterize using cell biological and biochemical approaches. We will extend our studies to other effector proteins and are currently using the CRISPR/Cas9 technology to engineer tomato plants with altered proteolytic profiles.
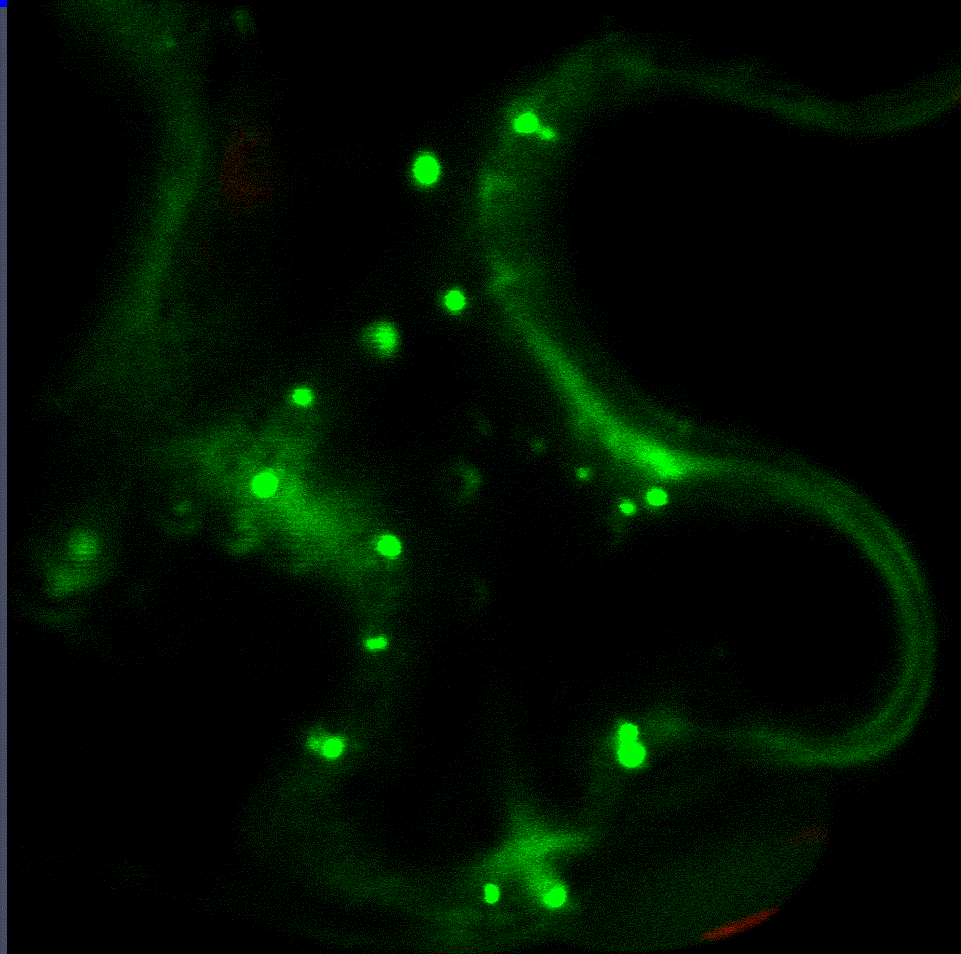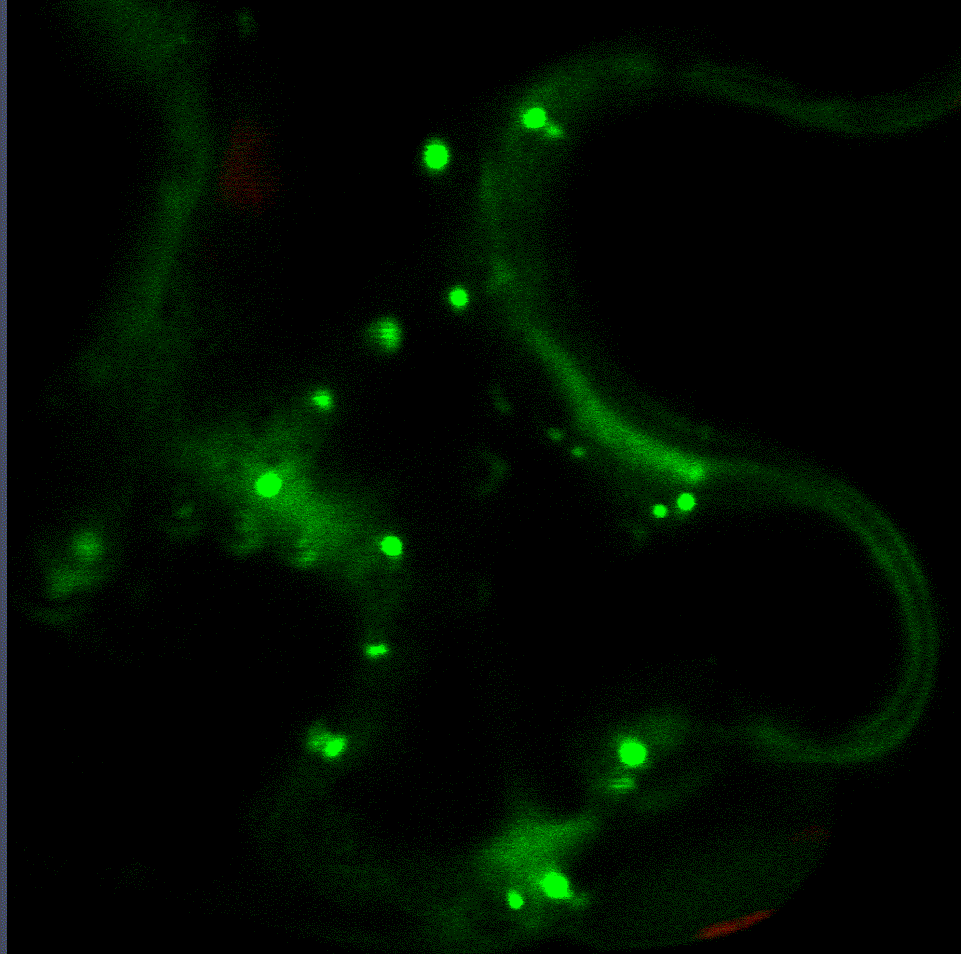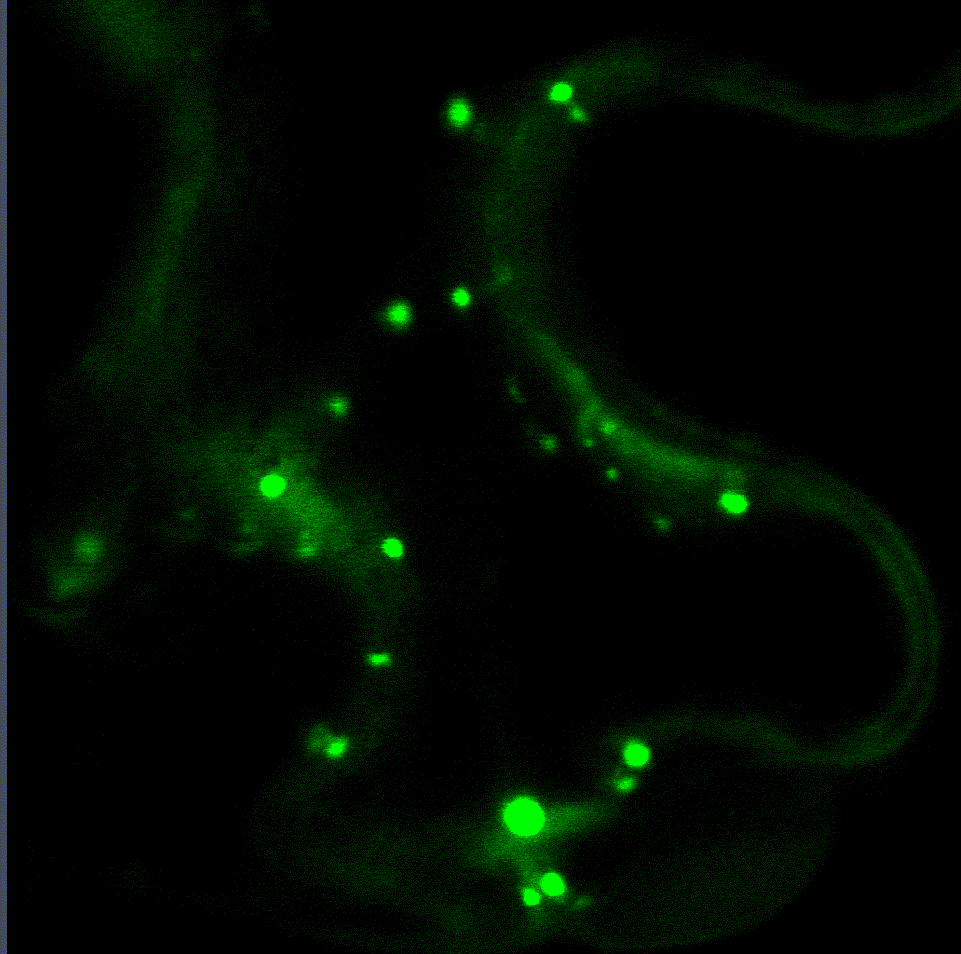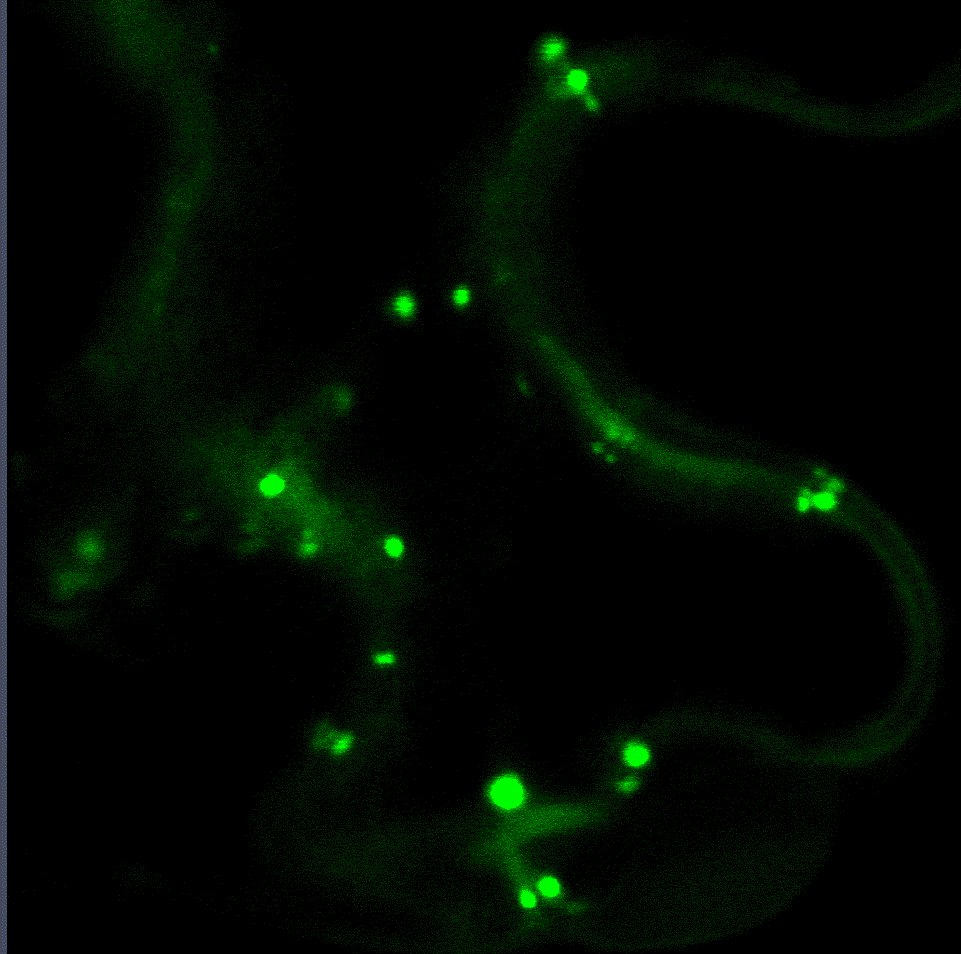
Regulatory fine-tuning of the proteasome is central to the outcome of plant-microbe interactions and seems to be a sensitive combination between down- and up-regulation of its activity. Therefore, we are currently analysing the mechanistic insights of how the proteasome is regulated on the transcriptional and post-translational level during plant immunity in a project funded through the CRC1101. Within this project we have identified two transcription factors that regulate proteasome gene expression during bacterial infection (Langin et al., unpublished). Interaction studies with both transcription factors revealed various proteins implicated in their degradation via the ERAD system and trafficking to the nucleus from the ER. Currently, we are looking into additional target genes of these transcription factors as they seem to mediate the trade-off between proteasome activation, growth and defence.

To protect themselves from these pathogens, plant immunity is efficiently regulated at the transcriptional and post-transcriptional level. The compartmentalization of mRNA into translationally repressed aggregates called processing bodies (PBs) is a key post-transcriptional regulatory process involved in development and stress responses. Preliminary results have shown that the bacterial plant pathogen Pseudomonas syringae (Pst) induces the formation of PB upon infection in an effector-dependent manner and that PB-defective Arabidopsis plants are more tolerant to Pst. This suggests that PBs are negative regulators of plant immunity that can be targeted by the bacterial effectors. The proposed project aims precisely at studying the role of PBs as post-transcriptional regulators of plant immunity and the ability of Pst effectors to modulate them. For this, a combination of genetic, biochemical, proteomic and cell biology approaches will be conducted to address: 1) the dynamics and roles of PBs, 2) the effector-mediated modulation of PB formation and 3) the interplay between PBs and autophagy, all in the context of a compatible plant-bacterium interaction.
In a more complete scenario, plants are constantly exposed to different pathogenic and beneficial microbes and hence it is crucial to include the bacteriome (microbiome) into this equation to obtain a holistic picture of the role of autophagy in plant-microbe interactions. The picture is getting even more complex if we look at cell-type specific autophagy response on the plant side. By studying how the microbiome and how different cell-types might shape autophagy and vice versa, we will get a deeper understanding of the role of autophagy in plant-microbe interactions. Thus, we will study in the ERC Starting Grant funded project DIVERSIPHAGY following questions and objectives:
In DIVERSIPHAGY, we will use a combination of state-of-the-art biochemical, proteomic, single-cell transcriptomics and cell-type specific reverse genetic approaches to decipher the role of autophagy in plant-microbe interactions. We aim to obtain a holistic view of how autophagy plays a role in plant-microbe interactions utilizing bacterial, genetic and cellular diversity with an emphasis on cell-type and organ-specific autophagy responses.